Pain Pathways Guide Researchers Toward a Safer Morphine
- Published1 Dec 2020
- Author Charlie Wood
- Source BrainFacts/SfN

Lurking in the shadows of the coronavirus pandemic, an unrelenting epidemic of opioid addiction rages on, fueled in no small part by a plentiful supply of illicit, powerful, and highly addictive synthetic opioids. The American Medical Association warned in May 2020 that the COVID-19 pandemic was driving overdoses as people couldn’t get the treatment they needed to overcome addiction. Underlying this tragedy is the fact that medicine currently lacks the tools to ameliorate pain without the risk of addiction.
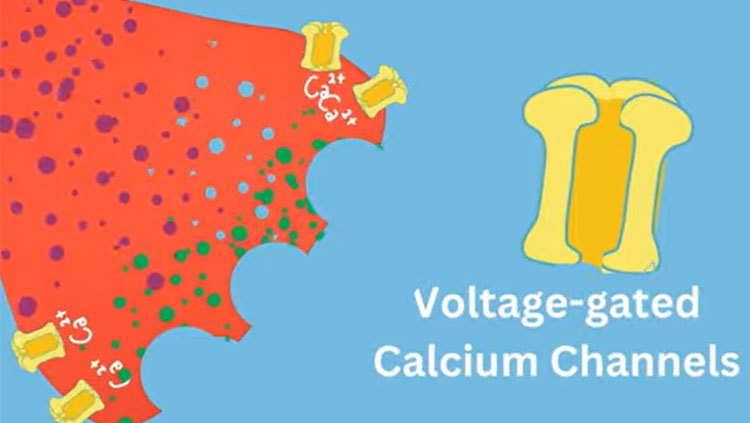
As scientists work to develop new effective, non-addictive therapies, they are focusing on the basic biological pathways that allow our bodies to sense pain. At the beginning, feeling pain relies on a set of proteins embedded in the cell membrane that open and close to control the rush of calcium into nerve cells. In fact, all our movements, thoughts, senses, and memories, our heartbeats, and our ability to speak, depend upon the function of these protein gates to propel signals through the neuron and trigger the of release neurotransmitters and pass the signal along to other neurons. Diane Lipscombe, a neuroscientist at Brown University, has devoted decades to teasing apart the myriad forms of these protein gates called calcium ion channels and exploring how they define a neuron’s activity.
“Calcium ion channels are at the beginning of the formation of signals in the nervous system,” says Lipscombe, the Society for Neuroscience’s past president, who has homed in on a particular type of calcium ion channels, the N-type voltage-gated calcium channels 2.2 and referred to in brief as the 2.2 channels, because this sub-type predominates in the nerve terminals of pain-sensing neurons.
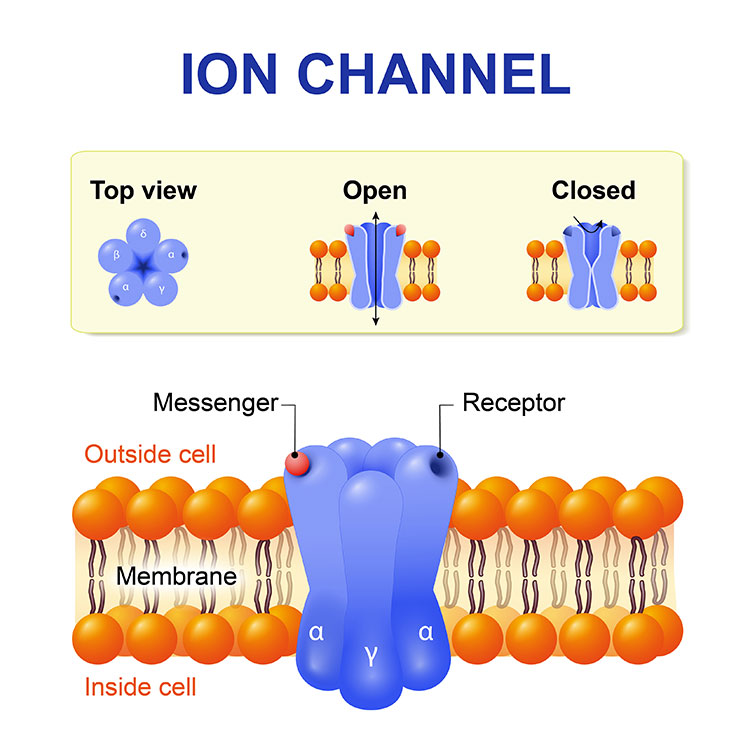
“In [pain receptors], we have identified a unique form of the 2.2 channel, and it has properties that just make sense,” she says. This type of channel is particularly responsive to opioids, for instance. “The calcium ion pathways across cells are therefore important sites for new drug development.” Since antiquity, we’ve relied on the blunt tool of opioids to quell pain.
Morphine, and all opioids, work in part by stopping 2.2 channels from opening. They successfully silence pain receptors but also cause addiction. A chronic pain medication called Prialt offers an analogous trade-off. It can interfere with pain-relaying calcium channels in the spinal cord but only if we accept an increased risk for hallucinations and confusion. An ideal pain therapy would shutter only the calcium ion channels in the body’s pain network without affecting those responsible for everything else.
“What you probably want to go for is something that’s somewhat preferential so it is effective at inhibiting pain signaling without shutting down the rest of the brain,” says Bruce Bean, a neurobiologist at Harvard.
That’s where Lipscombe’s work cataloging the enormous diversity of calcium ion channels comes in. Every 2.2 channel springs from identical genetic information housed in our DNA. Hundreds to potentially thousands of forms of this channel may exist in mammalian nervous systems. Each arising from cellular machinery knitting together that genetic information in a subtly different way.
It’s a strategy employed across the nervous system during neuronal differentiation, maturation, synapse formation, and more. Lipscombe suspects ensembles of specialized calcium ion channels may differ by brain area, with cerebellum channels tending to act a bit different from thalamus channels, for instance.
This is possible because of the way genetic information directs the production of proteins in our cells. DNA acts as a set of biological blueprints. In order to create the proteins, which do most of the work in the cell, the instructions held in the DNA double helix are transcribed into an RNA copy — a single strand of genetic information. From there, the cellular machinery reads the transcribed information, translates it, and assembles proteins.
This seemingly straightforward process contains a wrinkle known as alternative RNA splicing. The instructions for making many proteins don’t reside in one continuous string of DNA. Instead, coding regions of DNA called exons are interspersed with non-coding regions called introns. During the production of the RNA copy, or messenger RNA, the cell removes introns and splices the exons together to create a template for producing the proteins. Subtle variations in the way exons are spliced together can dictate how the proteins work.
This is notably true for the 2.2 channels involved in pain that Lipscombe studies. Her team recently discovered how the mechanism of alternative RNA splicing plays a role in the chronic pain pathology associated with peripheral nerve injury.
The team focused on the gene encoding a protein that is the functional core of the 2.2 channel called Cacna 1b. In pain-sensing neurons called nociceptors, one way of splicing together this genetic information gives rise to a 2.2 channel known as e37a, which enhances the pain-relieving activity of morphine. Combined in another way results in a channel known as e37b, which is less capable at quelling pain in response to morphine. What’s more, somehow following a nerve injury, the nociceptor ramps up production of the e37b form of the channel, reducing the ability of morphine to relieve pain.
It’s not anything that anyone would have thought about until her work started on this.
The team scoured publicly available databases looking for proteins capable of binding either DNA or messenger RNA at the point where the instructions drive the production of either morphine-sensitive e37a or morphine-resistant e37b. What they found was an abundant and ubiquitous protein that binds DNA. This protein is responsible for regulating how genes are expressed. It works basically by identifying small stretches of DNA sequences, binding to that DNA, and helping those regions stay open and available for the cellular machinery to get to work producing RNA and making protein.
The entire process of protein production can be ramped down or even silenced if the DNA is tagged with a molecule comprised of a carbon atom and three hydrogen atoms, known as a methyl group, that acts like a stop sign. Lipscombe and her team found that the morphine-sensitive e37a form predominated in nociceptor neurons when the associated DNA lacked methyl groups. However, when the e37a region of DNA included these methyl group stop signs, the e37b form appeared.
In a mouse model of nerve injury, the researchers discovered that while the e37a DNA started out unmethylated, over time the region was tagged with methyl groups, which stymied production of the e37a form and bolstered the production of the morphine-resistant e37b form. This mechanism could be at the root of neuropathic pain — pain caused by nerve injury.
Lipscombe suggests that a deeper understanding of how methylation patterns produce different calcium ion channels could someday lead to the development of more targeted pain killers. Such a compound could feature fewer, less severe side effects, such as intense addiction.
“It’s a novel direction,” Bean says. “It’s not anything that anyone would have thought about until her work started on this.” However, Bean maintains Lipscombe’s work is important beyond any drug development because it shows how complex ion channels can be.
Lipscombe agrees that exploring basic science has benefits outside the immediate development of new drugs. Just as geneticists cracked the code of how DNA serves as a blueprint, she hopes to continue drilling down into how the methylation drives identical genetic blueprints to develop site-specific variations. “If we can get at the code, then we can start to make predictions” about what kind of channel might get produced in a specific region of the nervous system, opening a new world of possibilities.
CONTENTPROVIDEDBY
BrainFacts/SfN
References
Lipscombe, D., & Andrade, A. (2015). Calcium Channel CaV Splice Isoforms - Tissue Specificity and Drug Action. Current Molecular Pharmacology, 8(1), 22–31. doi: 10.2174/1874467208666150507103215
Lipscombe, D., & Soto, E. J. L. (2019). Alternative splicing of neuronal genes: new mechanisms and new therapies. Current Opinion in Neurobiology, 57, 26–31. doi: 10.1016/j.conb.2018.12.013
Soto, E. J. L., & Lipscombe, D. (2020). Cell-specific exon methylation and CTCF binding in neurons regulates calcium ion channel splicing and function. doi: 10.7554/eLife.54879 eLife2020;9:e54879
(October 6, 2020). Issue brief: Reports of increases in opioid and other drug-related overdose and other concerns during COVID pandemic. American Medical Association. Retrieved from https://www.ama-assn.org/system/files/2020-10/issue-brief-increases-in-opioid-related-overdose.pdf
Thaler, C., Gray, A. C., & Lipscombe, D. (2004). Cumulative inactivation of N-type CaV2.2 calcium channels modified by alternative splicing. Proceedings of the National Academy of Sciences of the United States of America, 101(15), 5675. doi: 10.1073/pnas.0303402101
Striessnig, J. (2018). Getting a handle on CaV2.2 (N-type) voltage-gated Ca2+ channels. Proceedings of the National Academy of Sciences, 115(51), 12848. doi: 10.1073/pnas.1818608115