Two Novel Mouse Models to Visualize and Manipulate Microglia with High Selectivity
Dr. Chung is currently a postdoctoral associate at Baylor College of Medicine, Houston, TX. Follow them on Twitter @dchloechung.
Microglia, the brain-resident macrophages, have recently gained much interest in neuroscience due to their critical role in maintaining physiology as well as contributing to pathological changes in the brain. Such increased interest inevitably creates a greater need for in vivo tools that permit close examination of microglia-specific functions. Several mouse models targeting genes expressed by microglia or their progenitors, such as Tie2, Runx1, Sall1, or Cx3cr1, have already been used to deepen our understanding of microglia biology. Yet, selectivity for microglia was not always optimal, as it was often tricky to accurately distinguish microglia from other macrophages that have overlapping signatures of gene expression derived from the shared lineage. In their publication, Kaiser and Feng report two novel transgenic mouse models created by targeting a recently identified microglia-specific gene called Tmem119.
Recent cell-specific gene profiling studies have shown that Tmem119, a gene encoding transmembrane protein 119, is uniquely and robustly expressed in microglia. Although the function of TMEM119 is unclear, the high Tmem119 expression level specific to microglia suggested that this gene could be potentially targeted as a new tool to study microglia. Anti-TMEM119 antibodies are valuable to investigate microglial morphology and localization, but for some applications, including fluorescence activated cell sorting, live-imaging, or electrophysiology, it is advantageous to express a reporter. To this end, Kaiser and Feng designed novel mouse models to express either the fluorescent reporter protein EGFP or the recombinase CreET2 in TMEM119-expressing microglia, while keeping endogenous mouse Tmem119 expression intact to preserve its potentially crucial physiological function(s).
The authors utilized a CRISPR/Cas9-based knock-in approach to insert ribosome-skipping peptide porcine teschovirus-1 polyprotein (P2A), followed by EGFP or CreERT2, at the stop codon of endogenous murine Tmem119 (Figure 1A, 2A). This design approach was chosen to ensure that when TMEM119 is expressed in microglia, either EGFP or CreERT2 could be also expressed in the same cells. PCR analyses confirmed that this strategy successfully introduced the transgene into the Tmem119 locus in the novel Tmem119-EGFP and Tmem119-CreERT2 mouse models.
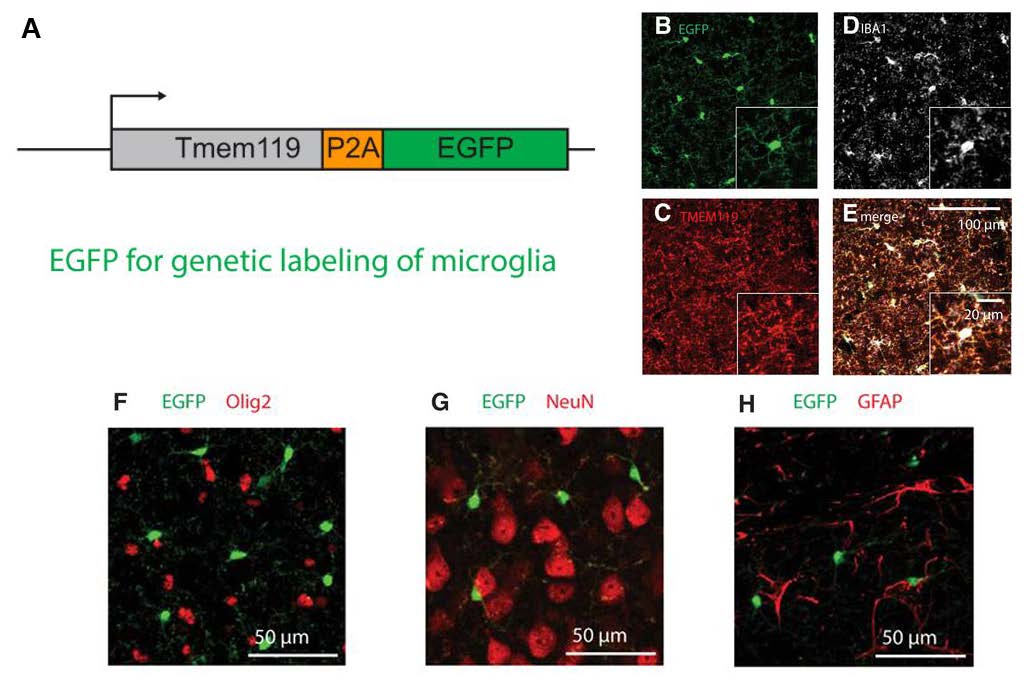
Characterization of Tmem119-EGFP mice demonstrated that endogenous murine Tmem119 levels as well as morphology of microglia remained highly intact. These observations indicate that the knock-in strategy had not drastically altered Tmem119 expression or general microglial properties. Subsequently, the authors determined that nearly all of the EGFP-expressing cells were also positive for the microglial marker IBA1 across various brain regions (Figure 1B-E). Antibodies to detect oligodendrocytes (Olig2), neurons (NeuN), astrocytes (glial fibrillary acidic protein; GFAP), and non-microglial macrophages (IBA1 immuno-reactivity combined with morphology) did not reveal colocalized expression in EGFP-positive cells (Figure 1F-H). These results indicate that EGFP expression is specific to TMEM119-expressing microglia, validating the fidelity of this mouse line. Excitingly, the sensitivity of microglial labeling was better than that of anti-TMEM119 antibodies as EGFP expression was first detected as early as postnatal day 1 (compared to postnatal 14 with antibody detection). This observation further highlights the value of this mouse model to study the early development of microglia.
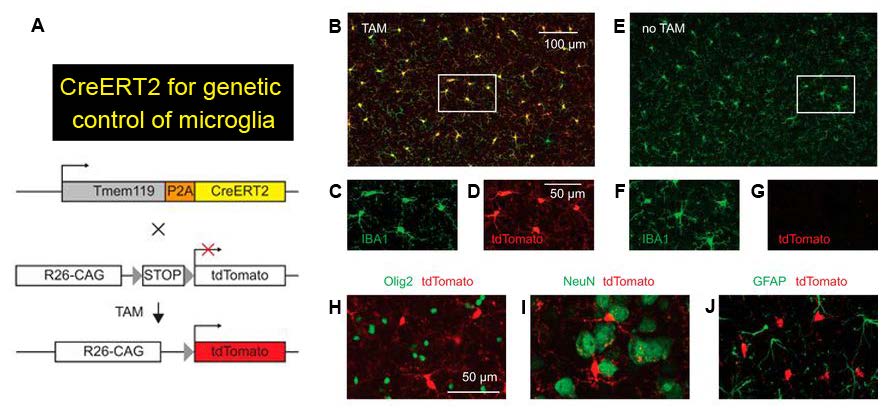
The authors characterized Tmem119-CreERT2 mice to check if the expression of CreERT2, a Cre recombinase that is activated by the estrogen receptor antagonist tamoxifen, is restricted to microglia. To do so, the authors crossed Tmem119-CreERT2 mice to Ai14 reporter mice that express the red fluorescent protein tdTomato in the presence of Cre activity (Figure 2A). Upon tamoxifen administration, tdTomato in the early postnatal and adult crossed mice selectively labeled microglia in a tamoxifen dose-dependent manner (Figure 2B-G). Importantly, oligodendrocytes, neurons, and astrocytes were not tdTomato-positive (Figure 2H-J). Almost all tdTomato-expressing cells were positive for the microglia marker IBA1, confirming that CreERT2 was faithfully expressed in microglia. However, unlike the Tmem119-EGFP line, ectopic Cre expression was unexpectedly detected in a small percentage of non-microglial cells such as IBA1-negative cells on the pia (Figure 3A-C) and cells of cortical blood vessels that are immuno-positive for the endothelial marker CD31 (Figure 3D-F). The authors reason that a very low level of Tmem119 expression in these non-microglial cells was still sufficient to induce Cre activity, while insufficient to express EGFP in the other novel mouse line.
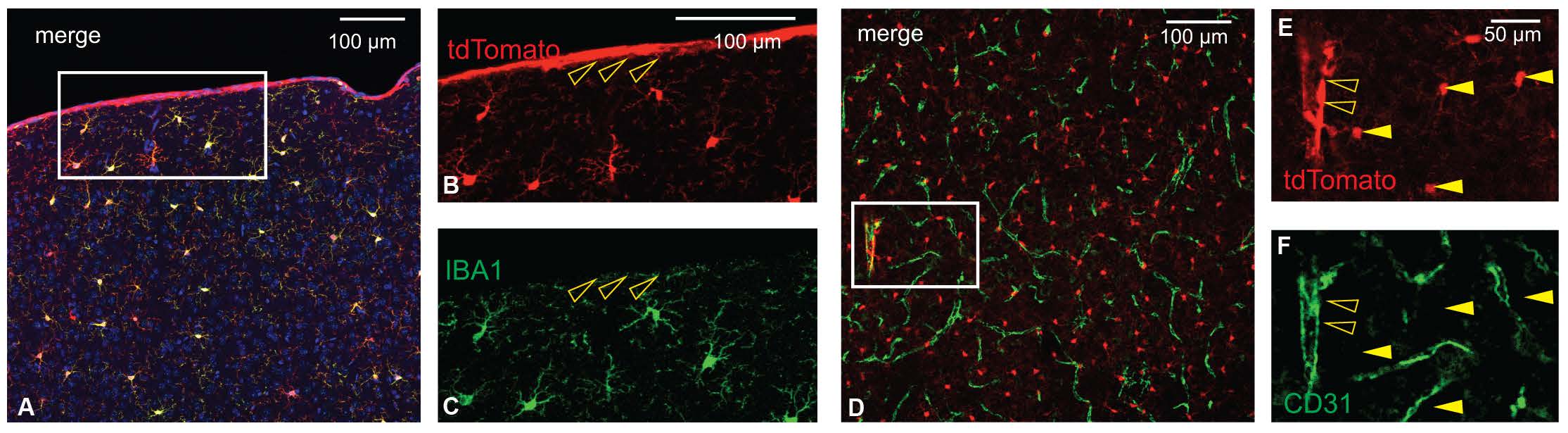
In summary, Kaiser and Feng have described two novel transgenic mouse models that can be used to specifically visualize or modulate microglial labeling in the brain. The newly generated Tmem119-EGFP and Tmem119-CreERT2 lines will bring value to the field with their ability to better distinguish microglia from other myeloid cells as well as their earlier timepoint of microglial labeling. As the significance of microglia continues to be discussed in a wide range of neurological diseases, many researchers will likely find these mouse models useful. The authors have already made both novel mouse lines available for research purposes via a public repository.
Read the full paper:
Tmem119-EGFP and Tmem119-CreERT2 Transgenic Mice for Labeling and Manipulating Microglia
Tobias Kaiser and Guoping Feng
FOLLOW US
POPULAR POSTS
TAGS
CATEGORIES


 RSS Feed
RSS Feed




