To Recombine or Not to Recombine
Nicole Amberg and Robert Beattie are currently post-doctoral fellows at the Institute of Science and Technology, You can follow on Twitter @amberg_nicole, @BeattieScience
Since its discovery, Cre-lox recombination technology has been one of the most widely adopted approaches for performing in vivo conditional or spatiotemporal genetic modifications (Gu et al., 1994). The Cre-recombinase enzyme catalyzes DNA recombination or excision events between specific target sequences termed loxP sites. By expressing the recombinase from a cell type- or tissue-specific promoter, biologists can address the function of distinct genes or signaling pathways in the development and maintenance of certain tissues. Temporal control of recombination events became possible with the advent of engineered recombinases fused to estrogen receptor (CreERT2). CreERT2 is bound by heat shock protein and remains inactive until the synthetic ligand tamoxifen is introduced (Feil et al., 1996).
However, some considerations should be taken into account when designing experiments that use the Cre-lox system to introduce a genetic modification with concurrent phenotypic assessment using fluorescent reporters. For example, a fluorescent reporter that is only expressed when a lox-stop-lox (LSL) cassette is removed, may not correlate 100% with the recombination of a second, separate floxed cassette in the same cell (Becher et al., 2018). This occurrence is thought to be due to each genomic region having a unique sensitivity to Cre activity, with some sites being more resistant to recombination than others.
In their publication, Dause & Kirby sought to quantitatively determine the rate of recombination of independent floxed alleles within individual cells. Since the Cre-lox technique is widely used in the field of adult neurogenesis, the authors targeted adult neural stem and progenitor cells (NSPCs) in the hippocampal subventricular zone and dentate gyrus using NestinCreERT2.
To determine whether recombination of one gene could be used to predict recombination in a second gene (Figure 1A), the authors first modeled the theoretical recombination rate for two LSL sequences. As shown in Figure 1B, a theoretically ideal scenario would result in a 100% overlap between the recombination of a fluorescent reporter gene (Gene R) and the recombination of a target gene (Gene T). By simulating the effects of variation in recombination efficiencies and suboptimal concordance rates for recombination of Gene R and Gene T, the model predicted that false positives and/or false negatives would emerge and lead to substantial error in data interpretation (Figure 1B).
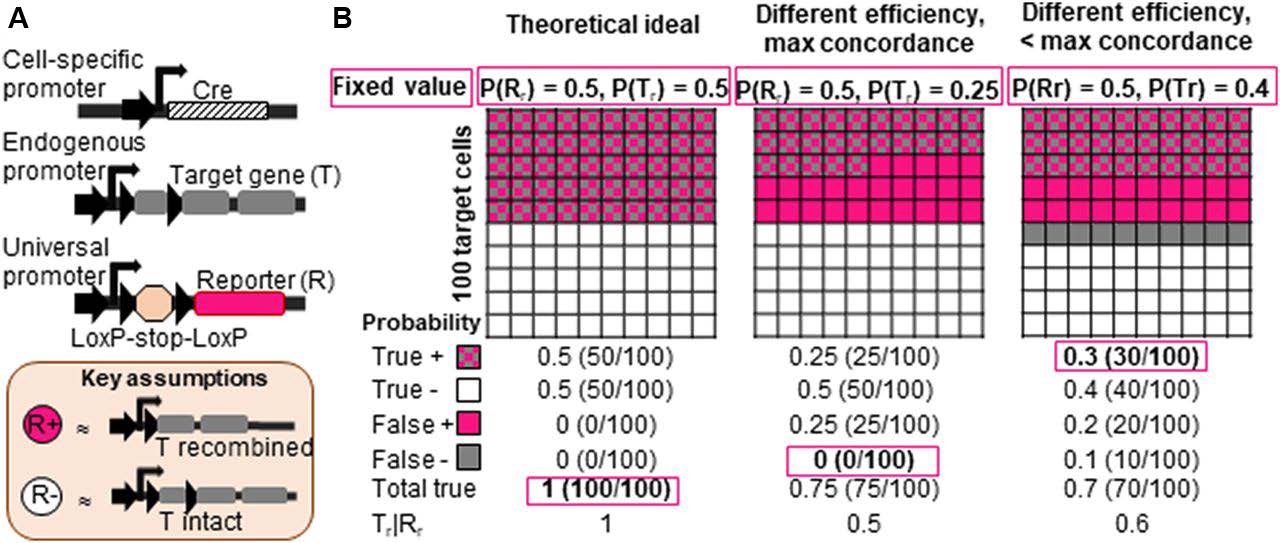
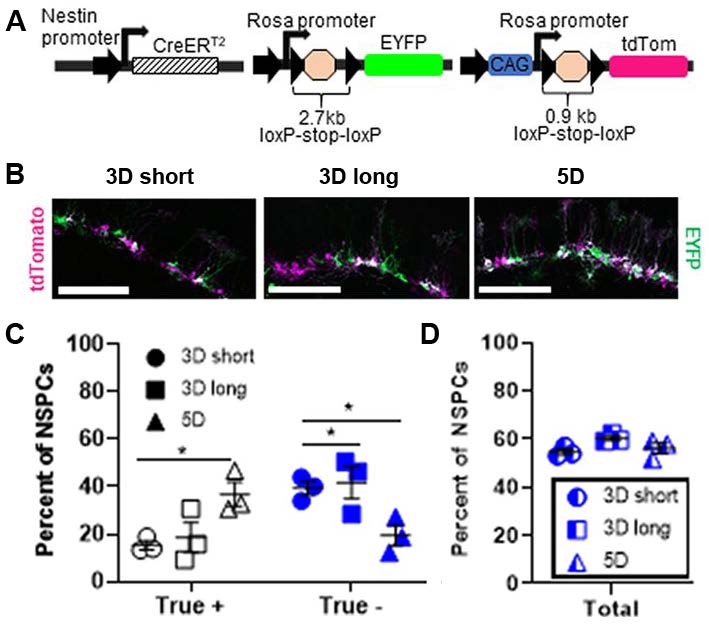
These results encouraged the authors to examine the relevance of their mathematical approach in a mouse model. Recombination of two distinct LSL-flanked fluorescent reporter constructs in adult NSPCs targeted with NestinCreERT2 was examined in male and female mice at 8–9 weeks of age. Both fluorescent reporter genes were targeted identically to the Rosa26 locus, thus rendering both sequences equally accessible to CreERT2. However, the tdTomato construct contains a CAG promoter and a 0.9 kb sequence between the loxP sites, whereas the EYFP construct does not have a CAG promoter and the distance between loxP sites is 2.7 kb (Figure 2A).
Ideally, transient expression of a recombinase would result in both LSL sequences being targeted with 100% efficiency, labeling all cells with tdTomato and EYFP. Surprisingly, the authors found that neither fluorescent reporter accurately predicted the expression of the other reporter, with only 50–60% overlap of the two reporters observed in NSPCs. By administering a variety of tamoxifen paradigms, they confirmed that this discordance in recombination frequency was independent of neural stem cell niche or tamoxifen administration protocol (Figure 2B-D).
The authors’ data suggest that it may not be reliable to assume that recombination of one gene is predictive of recombination of a second gene within single cells using tamoxifen-inducible Cre recombinases. Several factors may influence the recombination rate, including 1) the efficiency of the specific CreERT2 line, 2) individual locus accessibility, and 3) features of loxP-flanked constructs.
These results have implications for studies that rely on the Cre-lox system to make claims regarding cell autonomous function of genes in adult neurogenesis. While strong cell autonomous phenotypes might still be observed, these results of Dause & Kirby suggest that a labeled cell may be wild type, rather than recombined, due to non-congruent recombination. Thus, phenotypes at the population level may be ‘blurred’ by the noise of ‘false-positive’ and ‘false-negative’ events. This study provides inspiration for investigators to incorporate some of the many novel approaches and experimental designs currently available to biologists. For example, one way to ensure recombination of both reporter and gene of interest in the same cell could be to use single-cell RNA-seq to test if multiple floxed genes are present in each fluorescently labeled cell. For a histological read-out, reporters could be targeted to the same locus as the gene of interest, ensuring that readout and gene disruptions are the same (Becher et al., 2018). However, such an approach would require redesigning hundreds of published transgenic mice. Other ready-to-use, Cre-mediated genetic technology that provide a 100% correlation of reporter expression and target gene deletion is, for example, Mosaic Analysis with Dual Recombinase-mediated cassette exchange (MADR; Kim et al., 2019) or Mosaic Analysis with Double Markers (MADM; Zong et al., 2005). As such, MADM is a powerful dual-color labeling method that enables in vivo lineage tracing and concurrent genetic manipulation (of any candidate gene in principle) of individual progenitor cells and their progeny in mice (Beattie et al., 2017; Hippenmeyer et al., 2010; Laukoter et al., 2020; Zong et al., 2005).
Overall, the results of this study highlight the limitations of using inducible gene manipulation combined with LSL fluorescent reporters in adult NSPCs. These findings will have broad implications for future studies looking at cell autonomous gene function. This publication is an advance in the field as it provides a quantitative framework on the incidence of false-positive and false-negative recombination events and will thus encourage scientists to vigorously test the efficiency of their particular Cre-lox systems to mitigate potential systemic errors from experimental paradigms.
References:
- Beattie R, Postiglione MP, Burnett LE, Laukoter S, Streicher C, Pauler FM, et al. Mosaic Analysis with Double Markers Reveals Distinct Sequential Functions of Lgl1 in Neural Stem Cells.Neuron. 2017 May 3;94(3):517-533.e3.
- Becher B, Waisman A, Lu L-F. Conditional Gene-Targeting in Mice: Problems and Solutions.Immunity.2018 15;48(5):835–6.
- Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon P. Ligand-activated site-specific recombination in mice.Proc Natl Acad Sci USA. 1996 Oct 1;93(20):10887–90.
- Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science.1994 Jul 1;265(5168):103–6.
- Hippenmeyer S, Youn YH, Moon HM, Miyamichi K, Zong H, Wynshaw-Boris A, et al. Genetic mosaic dissection of Lis1 and Ndel1 in neuronal migration. Neuron.2010 Nov 18;68(4):695–709.
- Kim GB, Rincon Fernandez Pacheco D, Saxon D, Yang A, Sabet S, Dutra-Clarke M, et al. Rapid Generation of Somatic Mouse Mosaics with Locus-Specific, Stably Integrated Transgenic Elements. Cell. 2019 Sep 19;179(1):251-267.e24.
- Laukoter S, Beattie R, Pauler FM, Amberg N, Nakayama KI, Hippenmeyer S. Imprinted Cdkn1c genomic locus cell-autonomously promotes cell survival in cerebral cortex development.Nat Commun.2020 10;11(1):195.
- Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L. Mosaic analysis with double markers in mice.Cell.2005 May 6;121(3):479–92.
Read the full article:
Poor Concordance of Floxed Sequence Recombination in Single Neural Stem Cells: Implications for Cell Autonomous Studies
Tyler Joseph Dause and Elizabeth Diana Kirby
FOLLOW US
POPULAR POSTS
TAGS
CATEGORIES


 RSS Feed
RSS Feed




