Snapshots in Neuroscience: Hippocampal Neuron
This image has been selected to showcase the art that neuroscience research can create.
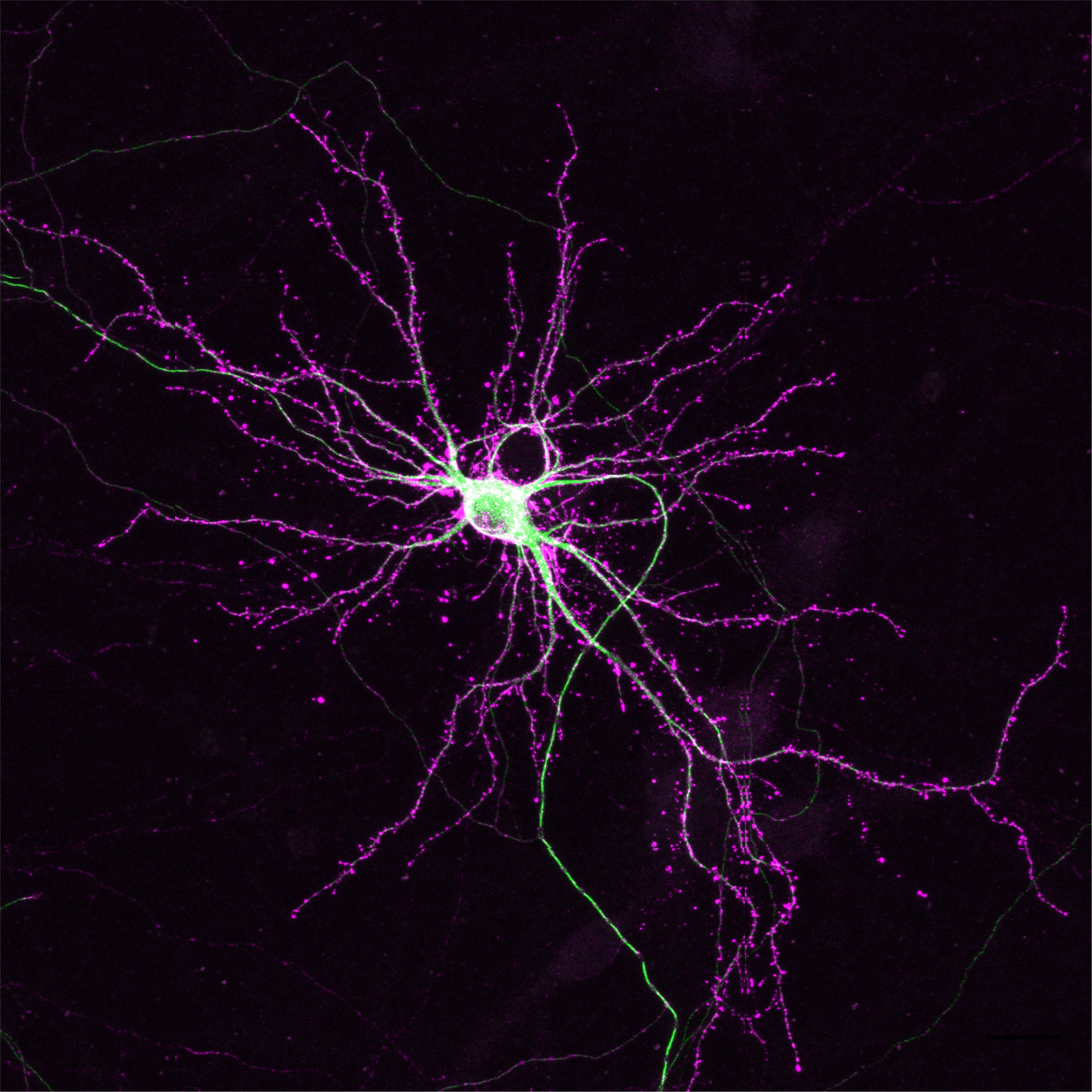
As described by Dr. de Jong and colleagues: The image shows a hippocampal neuron with endogenous labelling of the cytoskeleton proteins β-actin and β3 tubulin using CRISPR/Cas9. Over the recent years, several CRISPR/Cas9 knock-in techniques have been developed to accurately label proteins with bright fluorescent probes or epitope tags for accurate detection in live- and super resolution microscopy. These gene editing methods, however, were limited to editing a single gene, and technical hurdles prevented the generation of multiplex knock-ins. This study provides a solution for this problem: by editing genes one at a time with several days interval, reliable duplex gene editing can be performed in neuronal cells.
The neuron shown here first underwent gene editing of Tubb3 to create β3-tubulin-GFP (in green) and subsequently editing of Actb to generate HaloTag-β-actin (magenta). As expected for actin localization, the Halotag signal is strongly enriched in dendritic spines. GFP, on the other hand, is present in the soma and in neurites, with much reduced signal in dendritic spines; in line with β3-tubulin localization. This gene editing technology allows for accurate duplex labelling of neuronal proteins for the most demanding microscopy techniques.
Read the full article:
Duplex Labeling and Manipulation of Neuronal Proteins Using Sequential CRISPR/Cas9 Gene Editing
Wouter J. Droogers, Jelmer Willems, Harold D. MacGillavry, and Arthur P. H. de Jong
FOLLOW US
POPULAR POSTS
TAGS
CATEGORIES


 RSS Feed
RSS Feed




